Progress towards Kaggle-style workflows in Haskell
There’s been a lot of work in the Haskell ecosystem that has made it easier to write interactive Kaggle-like scripts. I’d like to showcase the synergy between 3 such tools: dataframe (my own creation), hasktorch, and IHaskell.
At a high level the intent of this post is to:
- get people excited about how far the ecosystem has come,
- checkpoint what the ecosystem currently looks like,
- identify areas of improvement in the workflow.
This post is a narrative companion to a hosted notebook, but with a little more context.
This work was done as part of the dataHaskell: an organization devoted to enabling reliable and reproducible machine learning using the Haskell programming language.
Special thanks to Jireh Tan for writing the code.
The iris classification problem
The Iris dataset is machine learning’s “Hello, World!” created by statistician Ronald Fisher in 1936. The dataset contains measurements of 150 iris flowers from three species:
- Setosa
- Versicolor
- Virginica
For each flower, we have four measurements (in centimetres):
- Sepal length and width (the green outer part)
- Petal length and width (the colourful inner part)
Our task is to predict the species of a flower given these four measurements.
Loading and Exploring the Data
We start by loading the data from a Parquet file using the DataFrame library:
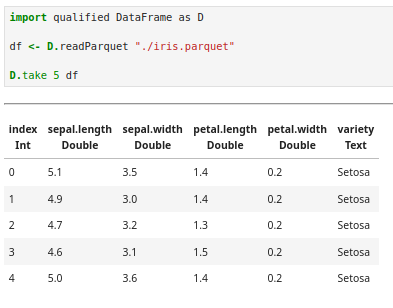
One immediate advantage of Haskell: the type system tells us we have no missing values. If any column contained nulls we’d see a Maybe prefix in front of that type. At least in this case, we don’t have to worry about deciding between imputing data and dropping it.
Exploring our data
Checking for balance
Before building any model, we need to know if our dataset is balanced. A severely imbalanced dataset (say, 140 Setosas and 5 each of the others) would make accurate prediction nearly impossible.
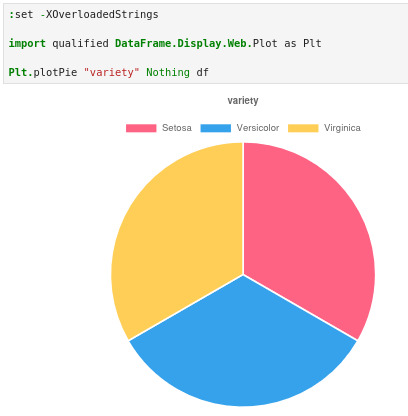
Great! A perfect balance with 50 flowers of each species. This means we don’t have to spend time on rebalancing tricks.
Some light analysis
Next, we compare feature magnitudes per class. We unintuitively do this in a stacked bar chart. But since our plot is interactive and allows us to exclude features at the click of a button we throw them all in there then click around to look at their individual differences.
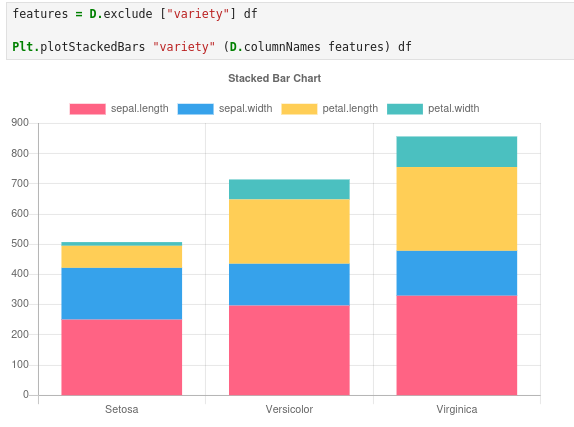
Our charts broadly suggest that Virginica > Versicolor > Setosa for most features, with one fun exception: Setosa has the largest sepal width.
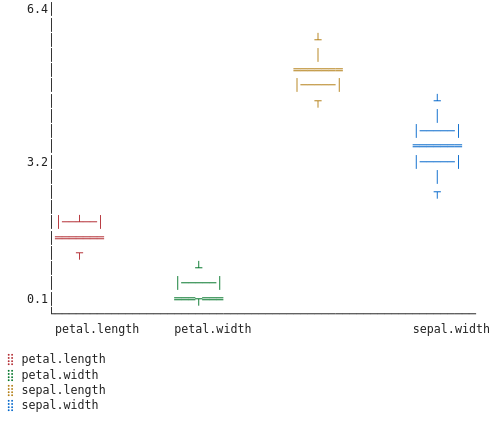
To get a sense of the spread of the features we can do box plots per variety and compare them:

Since we don’t have box plots implemented as web plots we fall back to granite’s ASCII plot.
For Setosa:
For Versicolor
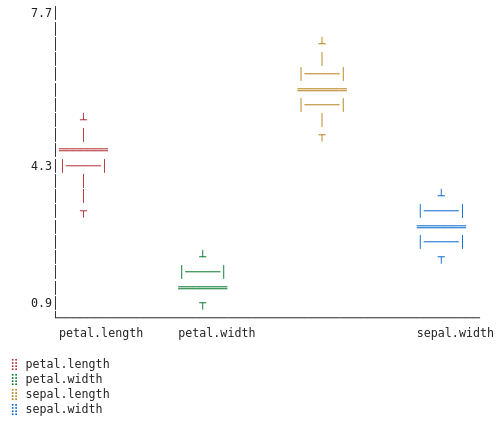
For Virginica:
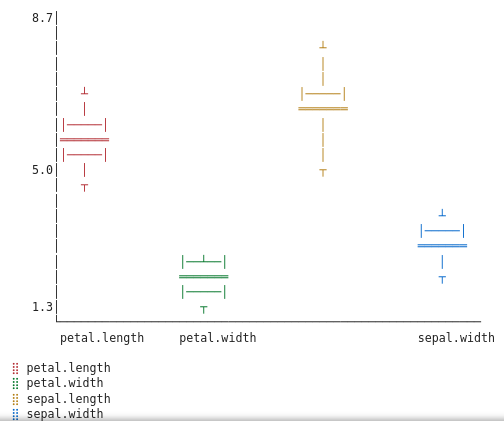
The box plots reveal that Setosas should be easy to identify—their petals are tiny with little variance. Versicolors and Virginicas overlap more, but we can still try to distinguish them using feature engineering.
Creating new features
From our exploration we have 5 promising feature candidates:
- sepal area
- sepal length to width ratio
- petal area
- petal length to width ratio
- sepal area / petal area
To derive these feature it’ll help to create typed references to the columns using template Haskell. This is the equivalent of doing a database migration against a schema definition. Except in this case we just freeze the types as they exist in the dataframe already.
:set -XTemplateHaskell
import qualified DataFrame.Functions as F
F.declareColumns df
import DataFrame ((|>))
engineered = df
|> D.derive "sepal_area" (sepal_length * sepal_width)
|> D.derive "sepal_ratio" (sepal_length / sepal_width)
|> D.derive "petal_area" (petal_length * petal_width)
|> D.derive "petal_ratio" (petal_length / petal_width)
|> D.derive "area_ratio" ((sepal_length * sepal_width) / (petal_length * petal_width))
After repeating the previous diagramming (see the full notebook) we decide we can get by with three features:
- petal_area,
- sepal_area, and,
- area_ratio
Creating a custom data type for our label
Here’s where Haskell’s type system shines. Instead of treating species as magic integers, we define them explicitly:
data Iris
= Setosa
| Versicolor
| Virginica
deriving (Eq, Show, Read, Ord, Enum)
The deriving clause automatically generates useful functions:
Eq: Allows us to compare Iris values for equalityShow: Converts Iris to a String (e.g., “Setosa”)Read: Converts a String to Iris (e.g., “Setosa” → Setosa)Ord: Allows ordering/sortingEnum: Lets us convert to/from integers (Setosa=0, Versicolor=1, Virginica=2)
We convert our string labels to this type:
import qualified Data.Text as T
modellingDf = D.select ["petal_area", "sepal_area", "area_ratio", "variety"] engineered
withTypedLabel =
modellingDf
|> D.derive
"variety"
(F.lift (fromEnum . read @Iris . T.unpack) variety)
Preparing our data for ML
For our model to generalise, we split the data: 70% for training, 30% for testing. Our random split function takes a seed and probability p. It return a tuple where (p * 100) % of the vakues are in the first element and the rest are in the second.
The random seed (42) ensures reproducibility which is crucial for debugging.
import qualified DataFrame.Hasktorch as DHT
import qualified System.Random as SysRand
import Control.Exception (throw)
let (trainDf, testDf) = D.randomSplit (SysRand.mkStdGen 42) 0.7 withTypedLabel
let trainFeaturesTr =
trainDf
|> D.exclude ["variety"]
|> DHT.toTensor
let testFeaturesTr =
testDf
|> D.exclude ["variety"]
|> DHT.toTensor
let trainLabels = either throw id (D.columnAsIntVector "variety" trainDf)
let testLabels = either throw id (D.columnAsIntVector "variety" testDf)
Since we are predicting one of many classes, our target should be a one-hot vector:
- 0 (Setosa) → [1.0, 0.0, 0.0]
- 1 (Versicolor) → [0.0, 1.0, 0.0]
- 2 (Virginica) → [0.0, 0.0, 1.0]
import qualified Torch as HT
let trainLabelsTr = HT.toType HT.Float (HT.oneHot 3 (HT.asTensor trainLabels))
let testLabelsTr = HT.toType HT.Float (HT.oneHot 3 (HT.asTensor $ testLabels))
The function application can be read from right to left:
- Make the label into a tensor
- Create a one-hot vector with three entries.
- Then finally convert each int to a float.
That’s it. Our data is ready for machine learning. Now we can pass it to Torch and use it to train a small multi-layer perceptron.
Training the model
Neural Network Architecture
We define our Multi-Layer Perceptron (MLP) architecture in two parts.
First, a specification that describes the shape of our network. Second, the actual model with its layers. Each layer is a Linear transformation (like nn.Linear in PyTorch).
:set -XDeriveGeneric
import qualified Torch as HT
import GHC.Generics (Generic)
data MLPSpec = MLPSpec
{ inputFeatures :: Int -- Number of input features (3 for our dataset)
, hiddenFeatures :: Int -- Number of neurons in hidden layer
, outputFeatures :: Int -- Number of output classes (3 species)
}
deriving (Show, Eq)
data MLP = MLP
{ l0 :: HT.Linear -- Input → Hidden layer
, l1 :: HT.Linear -- Hidden → Output layer
}
deriving (Generic, Show)
Network Architecture Diagram:
Input Layer (3) → Hidden Layer (8) → Output Layer (3)
--------------- ----------------- ----------------
sepal_area ReLU activation Softmax
petal_area (introduces (produces
area_ratio non-linearity) probabilities)
Setosa
Versicolor
Virginica
Making Our Model Trainable
We need to tell Hasktorch how to initialise our network with random weights.
This is similar to defining __init__() in a PyTorch nn.Module:
:set -XRecordWildCards
instance HT.Parameterized MLP
instance HT.Randomizable MLPSpec MLP where
sample MLPSpec{..} =
MLP
<$> HT.sample (HT.LinearSpec inputFeatures hiddenFeatures)
<*> HT.sample (HT.LinearSpec hiddenFeatures outputFeatures)
The <$> and <*> operators are Haskell’s way of working with random
initialisation. Think of this as: “Create an MLP by randomly sampling
weights for both layers.”
Forward Pass
This function defines how data flows through the network. It’s equivalent
to the forward() method in PyTorch. Read it from right to left (or
bottom to top in the chain):
mlp :: MLP -> HT.Tensor -> HT.Tensor
mlp MLP{..} =
HT.softmax (HT.Dim 1) -- 4. Apply softmax (probabilities sum to 1)
. HT.linear l1 -- 3. Apply second linear layer
. HT.relu -- 2. Apply ReLU activation
. HT.linear l0 -- 1. Apply first linear layer
Read this right-to-left (or bottom-to-top). The . operator composes functions.
In Python/PyTorch, this would look like:
def forward(self, x):
x = self.l0(x)
x = F.relu(x)
x = self.l1(x)
x = F.softmax(x, dim=1)
return x
The Haskell version is directly captures that we are composing a series of functions and using the output of one as the input to the other.
Training Loop
Training longer doesn’t always mean better. At some point, your model starts memorizing the training data instead of learning general patterns.
We detect this by watching the test set loss. As long as test loss keeps decreasing, the model is learning generalizable patterns. But if training loss keeps dropping while test loss starts climbing, we’re overfitting.
This is called early stopping, and it’s one of the simplest yet most effective regularisation techniques in machine learning.
The Training State
Our training loop needs to track more than just the current model. We maintain:
- Current model: Updated every iteration
- Best model: The weights that gave us the lowest test loss
- Best loss: What that test loss was
- Patience counter: How many checks since we last improved?
If we don’t improve for several consecutive checks (in our case 5 iterations), we give up and return the best model we found.
The loop structure might look unusual if you’re coming from PyTorch. Instead of a for loop with mutation, we use the very similar looking foldLoop which takes a number of iterations, an initial state and a function that evolves that state with each iteration. Each iteration receives the previous state and returns the new state. No hidden mutable variables, just explicit data flow.
This is eventually the sort of thing we should abstract behind a good machine learning library but for now it’s easy enough to implement.
import Control.Monad (when)
trainLoop ::
Int -> -- Number of epochs
(HT.Tensor, HT.Tensor) -> -- Training features and labels
(HT.Tensor, HT.Tensor) -> -- Test features and labels
MLP -> -- Initial model
IO MLP -- Returns trained model
trainLoop n trainingData testData initialModel = do
let initialState = makeInitialState initialModel
(_, bestModel, _, _) <- HT.foldLoop initialState n (trainingStep trainingData testData)
pure bestModel
where
-- Configuration constants
patience = 5
checkInterval = 500
learningRate = 1e-2
initialBestLoss = read @Float "Infinity"
-- Create initial training state
makeInitialState model = (model, model, initialBestLoss, 0)
-- Main training step for each iteration
trainingStep (features, labels) (testFeatures, testLabels) (model, bestModel, bestLoss, counter) i
| shouldStopEarly counter = pure (model, bestModel, bestLoss, counter)
| otherwise = do
-- Train and evaluate
(updatedModel, trainLoss) <- performTrainingStep model features labels
testLoss <- computeTestLoss updatedModel testFeatures testLabels
-- Log progress periodically
when (shouldLogProgress i) $
logTrainingProgress i trainLoss testLoss
-- Update best model tracking
let (newBest, newBestLoss, newCounter) =
updateBestModelTracking updatedModel testLoss bestModel bestLoss counter
pure (updatedModel, newBest, newBestLoss, newCounter)
-- Early stopping condition
shouldStopEarly counter = counter >= patience
-- Logging condition
shouldLogProgress i = i `mod` checkInterval == 0
-- Perform one training iteration
performTrainingStep model features labels = do
let predictions = mlp model features
loss = HT.binaryCrossEntropyLoss' labels predictions
(updatedModel, _) <- HT.runStep model HT.GD loss learningRate
pure (updatedModel, HT.asValue loss :: Float)
-- Evaluate model on test set
computeTestLoss model features labels =
let predictions = mlp model features
loss = HT.binaryCrossEntropyLoss' labels predictions
in pure (HT.asValue loss :: Float)
-- Print training progress
logTrainingProgress iteration trainLoss testLoss =
putStrLn $ "Iteration: " ++ show iteration
++ " | Training Set Loss: " ++ show trainLoss
++ " | Test Set Loss: " ++ show testLoss
-- Update best model if test loss improved, otherwise increment patience counter
updateBestModelTracking currentModel currentLoss bestModel bestLoss counter =
if currentLoss < bestLoss
then (currentModel, currentLoss, 0) -- New best: reset counter
else (bestModel, bestLoss, counter + 1) -- No improvement: increment counter
Running the training loop
We bring everything together by running our training loop with a model intialized from our MLPSpec.
initialModel <- HT.sample $ MLPSpec 3 8 3
trainedModel <-
trainLoop
10000
(trainFeaturesTr, trainLabelsTr)
(testFeaturesTr, testLabelsTr)
initialModel
putStrLn "Your model weights are given as follows: "
print trainedModel
Evaluating the model
The notebook creates some helps to evaluate to the model. We’ll skip these details since most of the code is vanilla Haskell.
And for our results. Training metrics:
------------------------------------------
index | variety | precision | recall
------|------------|-----------|----------
Int | Iris | Float | Float
------|------------|-----------|----------
0 | Setosa | 1.0 | 1.0
1 | Versicolor | 0.9677419 | 0.9375
2 | Virginica | 0.9459459 | 0.9722222
Test metrics:
------------------------------------------
index | variety | precision | recall
------|------------|-----------|----------
Int | Iris | Float | Float
------|------------|-----------|----------
0 | Setosa | 1.0 | 0.9285714
1 | Versicolor | 0.8888889 | 0.8888889
2 | Virginica | 0.8666667 | 0.9285714
Conclusion
This post demonstrates building a complete multiclass classification system in Haskell from scratch. We tackle the classic Iris dataset. The journey covers data loading, exploratory analysis, feature engineering, typed data modelling, network architecture design, training with early stopping, and evaluation.
The tools for a robust data science journey are scattered in the Haskell ecosystem. The hope is that we can unify them and along the way create ergonomic and safe APIs for data science.
Wanna join in this work?
Come over to the dataHaskell Discord to help shape the future of data science and Haskell?
The downloaded ipynb for this post can be found here.